决策树 Decision_trees - Ex 4: Understanding the decision tree structure
决策树范例四: Understanding the decision tree structure
http://scikit-learn.org/stable/auto_examples/tree/plot_unveil_tree_structure.html
范例目的
此范例主要在进一步探讨决策树内部的结构,分析以获得特征与目标之间的关係,并进而进行预测。
- 当每个节点的分支最多只有两个称之为二元树结构。
- 判断每个深度的节点是否为叶,在二元树中若该节点为判断的最后一层称之为叶。
- 利用
decision_path获得决策路径的资讯。 - 利用
apply得到预测结果,也就是决策树最后抵达的叶。 - 建立完成后的规则变能用来预测。
- 一组多个样本可以寻得其中共同的决策路径。
(一)引入函式库及测试资料
引入函式资料库
load_iris引入鸢尾花资料库。
from sklearn.model_selection import train_test_splitfrom sklearn.datasets import load_irisfrom sklearn.tree import DecisionTreeClassifier
建立训练、测试集及决策树分类器
- X (特征资料) 以及 y (目标资料)。
train_test_split(X, y, random_state)将资料随机分为测试集及训练集。
X为特征资料集、y为目标资料集,random_state随机数生成器。DecisionTreeClassifier(max_leaf_nodes, random_state)建立决策树分类器。max_leaf_nodes节点为叶的最大数目,random_state若存在则为随机数生成器,若不存在则使用np.random。fit(X, y)用做训练,X为训练用特征资料,y为目标资料。
iris = load_iris()X = iris.datay = iris.targetX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)estimator = DecisionTreeClassifier(max_leaf_nodes=3, random_state=0)estimator.fit(X_train, y_train)
(二) 决策树结构探讨
在DecisionTreeClassifier 中有个属性 tree_,储存了整个树的结构。
二元树被表示为多个平行的矩阵,每个矩阵的第i个元素储存着关于节点”i”的信息,节点0代表树的根。
需要注意的是,有些矩阵只适用于有分支的节点,在这种情况下,其他类型的节点的值是任意的。
上述所说的矩阵包含了:
node_count:总共的节点个数。children_left:节点左边的节点的ID,”-1”代表该节点底下已无分支。children_righ:节点右边的节点的ID,”-1”代表该节点底下已无分支。feature:使节点产生分支的特征,”-2”代表该节点底下已无分支。threshold:节点的阀值。若距离不超过 threshold ,则边的两端就视作同一个群集。
n_nodes = estimator.tree_.node_countchildren_left = estimator.tree_.children_leftchildren_right = estimator.tree_.children_rightfeature = estimator.tree_.featurethreshold = estimator.tree_.threshold
以下为各矩阵的内容
n_nodes = 5children_left [ 1 -1 3 -1 -1]children_right [ 2 -1 4 -1 -1]feature [ 3 -2 2 -2 -2]threshold [ 0.80000001 -2. 4.94999981 -2. -2. ]
二元树的结构所通过的各个属性是可以被计算的,例如每个节点的深度以及是否为树的最底层。
node_depth:节点在决策树中的深度(层)。is_leaves:该节点是否为决策树的最底层(叶)。stack:存放尚未判断是否达决策树底层的节点资讯。
将stack的一组节点资讯pop出来,判断该节点的左边节点ID是否等于右边节点ID。
若不相同分别将左右节点的资讯加入stack中,若相同则该节点已达底层is_leaves设为True。
node_depth = np.zeros(shape=n_nodes)is_leaves = np.zeros(shape=n_nodes, dtype=bool)stack = [(0, -1)] #initialwhile len(stack) > 0:node_id, parent_depth = stack.pop()node_depth[node_id] = parent_depth + 1# If we have a test nodeif (children_left[node_id] != children_right[node_id]):stack.append((children_left[node_id], parent_depth + 1))stack.append((children_right[node_id], parent_depth + 1))else:is_leaves[node_id] = True
执行过程
stack len 1node_id 0 parent_depth -1node_depth [ 0. 0. 0. 0. 0.]stack [(1, 0), (2, 0)]stack len 2node_id 2 parent_depth 0node_depth [ 0. 0. 1. 0. 0.]stack [(1, 0), (3, 1), (4, 1)]stack len 3node_id 4 parent_depth 1node_depth [ 0. 0. 1. 0. 2.]stack [(1, 0), (3, 1)]stack len 2node_id 3 parent_depth 1node_depth [ 0. 0. 1. 2. 2.]stack [(1, 0)]stack len 1node_id 1 parent_depth 0node_depth [ 0. 1. 1. 2. 2.]stack []
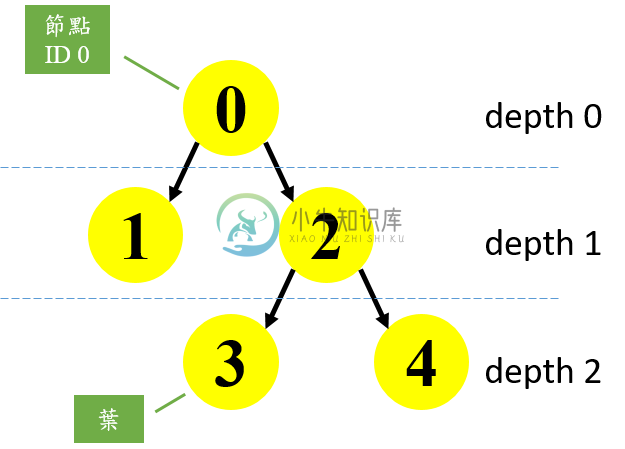
下面这个部分是以程式的方式印出决策树结构,这个决策树共有5个节点。
若遇到的是test node则用阀值决定该往哪个节点前进,直到走到叶为止。
print("The binary tree structure has %s nodes and has ""the following tree structure:"% n_nodes)for i in range(n_nodes):if is_leaves[i]:print("%snode=%s leaf node." % (node_depth[i] * "\t", i)) #"\t"缩排else:print("%snode=%s test node: go to node %s if X[:, %s] <= %s else to ""node %s."% (node_depth[i] * "\t",i,children_left[i],feature[i],threshold[i],children_right[i],))
执行结果
The binary tree structure has 5 nodes and has the following tree structure:node=0 test node: go to node 1 if X[:, 3] <= 0.800000011921 else to node 2.node=1 leaf node.node=2 test node: go to node 3 if X[:, 2] <= 4.94999980927 else to node 4.node=3 leaf node.node=4 leaf node.
接下来要来探索每个样本的决策路径,利用decision_path方法可以让我们得到这些资讯,apply存放所有sample最后抵达哪个叶。
以第0笔样本当作范例,indices存放每个样本经过的节点,indptr存放每个样本存放节点的位置,node_index中存放了第0笔样本所经过的节点ID。
node_indicator = estimator.decision_path(X_test)# Similarly, we can also have the leaves ids reached by each sample.leave_id = estimator.apply(X_test)# Now, it's possible to get the tests that were used to predict a sample or# a group of samples. First, let's make it for the sample.sample_id = 0node_index = node_indicator.indices[node_indicator.indptr[sample_id]:node_indicator.indptr[sample_id + 1]]print('node_index', node_index)print('Rules used to predict sample %s: ' % sample_id)for node_id in node_index:if leave_id[sample_id] != node_id:continueif (X_test[sample_id, feature[node_id]] <= threshold[node_id]):threshold_sign = "<="else:threshold_sign = ">"print("decision id node %s : (X[%s, %s] (= %s) %s %s)"% (node_id,sample_id,feature[node_id],X_test[i, feature[node_id]],threshold_sign,threshold[node_id]))
执行结果
node_index [0 2 4]Rules used to predict sample 0:decision id node 4 : (X[0, -2] (= 1.5) > -2.0)
接下来是探讨多个样本,是否有经过相同的节点。
以样本0、1当作范例,node_indicator.toarray()存放多个矩阵0代表没有经过该节点,1代表经过该节点。common_nodes中存放true与false,若同一个节点相加的值等于输入样本的各树,则代表该节点都有被经过。
# For a group of samples, we have the following common node.sample_ids = [0, 1]common_nodes = (node_indicator.toarray()[sample_ids].sum(axis=0) ==len(sample_ids))print('node_indicator',node_indicator.toarray()[sample_ids])print('common_nodes',common_nodes)common_node_id = np.arange(n_nodes)[common_nodes]print('common_node_id',common_node_id)print("\nThe following samples %s share the node %s in the tree"% (sample_ids, common_node_id))print("It is %s %% of all nodes." % (100 * len(common_node_id) / n_nodes,))
执行结果
node_indicator [[1 0 1 0 1][1 0 1 1 0]]common_nodes [ True False True False False]common_node_id [0 2]The following samples [0, 1] share the node [0 2] in the treeIt is 40.0 % of all nodes.
(三)完整程式码
import numpy as npfrom sklearn.model_selection import train_test_splitfrom sklearn.datasets import load_irisfrom sklearn.tree import DecisionTreeClassifieriris = load_iris()X = iris.datay = iris.targetX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)estimator = DecisionTreeClassifier(max_leaf_nodes=3, random_state=0)estimator.fit(X_train, y_train)# The decision estimator has an attribute called tree_ which stores the entire# tree structure and allows access to low level attributes. The binary tree# tree_ is represented as a number of parallel arrays. The i-th element of each# array holds information about the node `i`. Node 0 is the tree's root. NOTE:# Some of the arrays only apply to either leaves or split nodes, resp. In this# case the values of nodes of the other type are arbitrary!## Among those arrays, we have:# - left_child, id of the left child of the node# - right_child, id of the right child of the node# - feature, feature used for splitting the node# - threshold, threshold value at the node## Using those arrays, we can parse the tree structure:n_nodes = estimator.tree_.node_countchildren_left = estimator.tree_.children_leftchildren_right = estimator.tree_.children_rightfeature = estimator.tree_.featurethreshold = estimator.tree_.threshold# The tree structure can be traversed to compute various properties such# as the depth of each node and whether or not it is a leaf.node_depth = np.zeros(shape=n_nodes)is_leaves = np.zeros(shape=n_nodes, dtype=bool)stack = [(0, -1)] # seed is the root node id and its parent depthwhile len(stack) > 0:node_id, parent_depth = stack.pop()node_depth[node_id] = parent_depth + 1# If we have a test nodeif (children_left[node_id] != children_right[node_id]):stack.append((children_left[node_id], parent_depth + 1))stack.append((children_right[node_id], parent_depth + 1))else:is_leaves[node_id] = Trueprint("The binary tree structure has %s nodes and has ""the following tree structure:"% n_nodes)for i in range(n_nodes):if is_leaves[i]:print("%snode=%s leaf node." % (node_depth[i] * "\t", i))else:print("%snode=%s test node: go to node %s if X[:, %s] <= %ss else to ""node %s."% (node_depth[i] * "\t",i,children_left[i],feature[i],threshold[i],children_right[i],))print()# First let's retrieve the decision path of each sample. The decision_path# method allows to retrieve the node indicator functions. A non zero element of# indicator matrix at the position (i, j) indicates that the sample i goes# through the node j.node_indicator = estimator.decision_path(X_test)# Similarly, we can also have the leaves ids reached by each sample.leave_id = estimator.apply(X_test)# Now, it's possible to get the tests that were used to predict a sample or# a group of samples. First, let's make it for the sample.sample_id = 0node_index = node_indicator.indices[node_indicator.indptr[sample_id]:node_indicator.indptr[sample_id + 1]]print('Rules used to predict sample %s: ' % sample_id)for node_id in node_index:if leave_id[sample_id] != node_id:continueif (X_test[sample_id, feature[node_id]] <= threshold[node_id]):threshold_sign = "<="else:threshold_sign = ">"print("decision id node %s : (X[%s, %s] (= %s) %s %s)"% (node_id,sample_id,feature[node_id],X_test[i, feature[node_id]],threshold_sign,threshold[node_id]))# For a group of samples, we have the following common node.sample_ids = [0, 1]common_nodes = (node_indicator.toarray()[sample_ids].sum(axis=0) ==len(sample_ids))common_node_id = np.arange(n_nodes)[common_nodes]print("\nThe following samples %s share the node %s in the tree"% (sample_ids, common_node_id))print("It is %s %% of all nodes." % (100 * len(common_node_id) / n_nodes,))

