Encode: Multidimensional encoding of brain connectomes
About
This software implements a framework to encode structural brain connectomes into multidimensional arrays. These arrays are commonly referred to as tensors. Encoding Connectomes provides an agile framework for computing over connectome edges and nodes efficiently. We provide several examples of operations that can be performed using the framework.
One major application of the tensor encoding is the implementation of the Linear Fascicle Evaluation method, in short LiFE. The tensor encoding method allows implementing LiFE with a dramatic reduction in storage requirements, up to 40x compression factors. Furthermore, connectome encoding allows performing multiple computational neuroanatomy operations such as tract-dissections, virtual lesions, and connectivity estimates very efficiently using machine-friendly array operators.
We provide demos to explain how to:(1) Load and encode diffusion-weighted data and tractography models of white matter fascicles, as well as perform multidimensional arrays operations.(2) Build and optimize a Linear Fascicle Evaluation model.(3) Perform neuroanatomical segmentation, computational neuroanatomy operations, and virtual lesions using the connectome encoding framework.(4) Reproduce some of the figures of an article describing the method implemented in this toolbox: Caiafa, C.F., Pestilli, F. Multidimensional encoding of brain connectomes. Sci Rep 7, 11491 (2017). https://doi.org/10.1038/s41598-017-09250-w
Application.
- Encoding of brain conenctome and associated phenotypes into multidimensional arrays.
- Evaluate the evidence supporting white-matter connectomes generated using magnetic resonance diffusion-weighted imaging and computational tractography .
- Perform statistical inference on white-matter connectomes: Compare white-matter connectomes, show the evidence for white-matter tracts and connections between brain areas.
License.
Copyright (2017), Cesar Caiafa, ccaiafa@gmail.com and Franco Pestilli, frakkopesto@gmail.com
Stable code release.
How to cite the software.
Funding.
Installation.
- Download (https://github.com/brain-life/encode).
- Start MatLab.
- Add repository to the matlab search path.
Dependencies.
Getting started.
1. Download the repository.
- Download the Encode repository from the TAR/ZIP files linked here.
- UNZIP/UNTAR the file.
- Add the encode folder to your matlab search path. To do so in the MatLab prompt type:
>> addpath(genpath('/my/path/to/the/encode/folder/'))
2. Download the vistasoft repository.
- Download the VISTASOFT repository from the TAR/ZIP files linked here.
- UNZIP/UNTAR the file.
- Add the VISTASOFT folder to your matlab search path. To do so in the MatLab prompt type:
>> addpath(genpath('/my/path/to/the/VISTASOFT/folder/'))
3. Download the MBA repository.
- Download the MBA repository from the TAR/ZIP files linked here.
- UNZIP/UNTAR the file.
- Add the MBA folder to your matlab search path. To do so in the MatLab prompt type:
>> addpath(genpath('/my/path/to/the/MBA/folder/'))
4. Download the Demo Datasets.
- Download the demo datasets from the repository doi:10.5967/K8X63JTX.
- UNTAR the main file Demo_Data_for_Multidimensional_Encoding_of_Brain_Connectomes.tar.gz
- Go inside the folder Demo_Data_for_Multidimensional_Encoding_of_Brain_Connectomes/ and UNZIP the following files: Figs_data.zip, HCP3T.zip, HCP7T, and STN. You can deleted the original .zip files once they are unziped.
- The structures of files and foldes under the main folder should looks like as follows
- feDemoDataPath.m
- Figs_data/
- HCP3T/
- HCP7T/
- README.txt
- STN/
- Add the main data folder (Demo_Data_for_Multidimensional_Encoding_of_Brain_Connectomes/) to your matlab search path. To do so in the MatLab prompt type:
>> addpath(genpath('/my/path/to/the/Demo_Data_for_Multidimensional_Encoding_of_Brain_Connectomes/'))
5. Run the demo_connectome_encoding code.
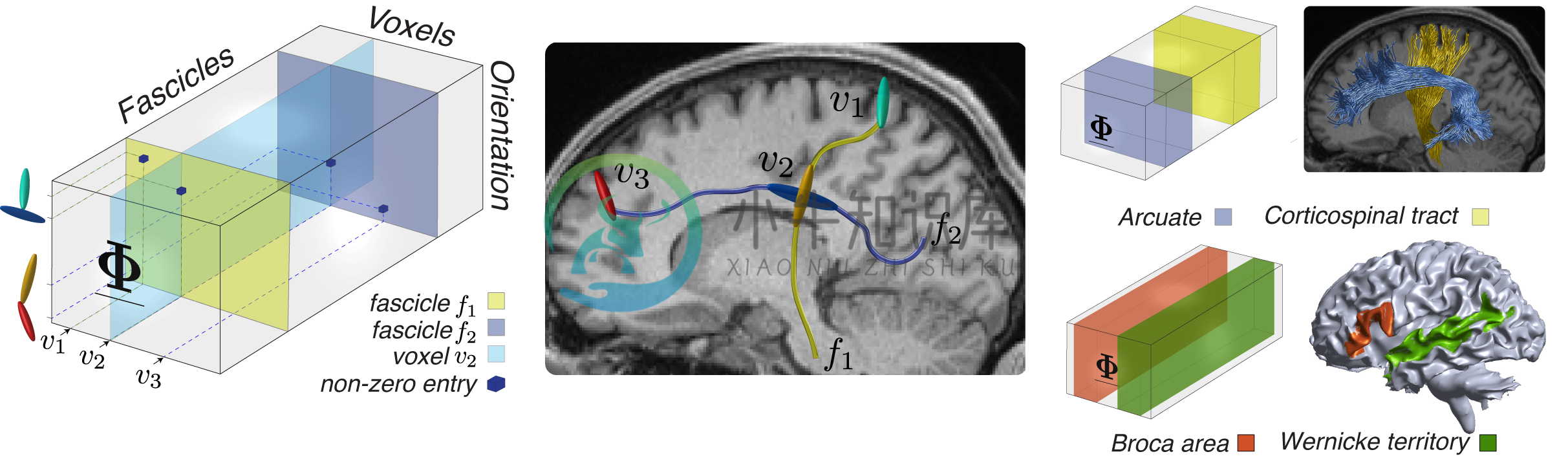
Here you will learn about creating the tensor representation of a connectoms and perform basic operations such as identifying fascicles having a particular spatial orientation in a small voxel area.
>> demo_connectome_encoding.m
6. Run the demo_connectome_data_comparison code.
This code reproduce Fig. 3 of the paper "Multidimensional encoding of brain connectomes", by C. Caiafa and F. Pestilli.
>> demo_connectome_data_comparison.m
7. Run the demo_virtual_lesion code.
This code allows you to compute virtual lesions on a particular brain dataset and visualize particular major tracts together with their path-neighborhood, i.e. fascicles sharing same voxels.
>> demo_virtual_lesion.m
8. Run the demo_LiFE code.
This code allows you to compute compute the fascicles weights for two different tractography methods, probabilistic and deterministic tractographies, on a same brain. This is similar to the original LiFE demo in https://github.com/francopestilli/life but here a full brain dataset is used. The optimization (fitting fascicles weights) runs in about 3 hours on a modern Intel processor with 8GB of RAM. This code has been tested with MatLab 2015b on Ubuntu 15+ and Mac OSX 10.11.
>> demo_LiFE.m
9. Description of the output data structure (fe.mat)
fe fields
• fe.name: [text], customized name of the structure.
• fe.type: [text], type description.
• fe.life: [1 × 1 struct], results of the LiFE method, see fe.life fields description below.
• fe.fg: [1 × 1 struct], input fiber group (connectome) information, see fe.fg fields description below.
• fe.roi: [1 × 1 struct], input fiber group (connectome) information, see fe.roi fields description below.
• fe.path: [1 × 1 struct], paths to input data (connectome, dMRI data, etc.).
• fe.rep: [1×1 struct], dMRI data for a repeated measurement if available.
fe.life fields
• fe.life.M.Phi: [Na × Nv × Nf sptensor], sparse array Φ encoding the connectome.
• fe.life.M.Nphi: Discretization number in azimuth, default = 360.
• fe.life.M.Ntheta: Discretization number in elevation, default = 360.
• fe.life.M.orient: [3 × Na double], matrix containing in its columns the orientations for each Dictionary element.
• fe.life.M.DictSig: [Nθ × Na double], Dictionary matrix containing in its columns the canonical diffusion kernel (demeaned) at different orienta- tions.
• fe.life.xform.img2acpc: [4 × 4 double], Image to ACPC affine trans- form.
• fe.life.xform.acpc2img: [4 × 4 double], ACPC to image affine trans- form.
• fe.life.fibers: not used.
• fe.life.diffusion_signal_img: [Nv × Nθ double], dMRI data.
• fe.life.diffusion_S0_img: [Nv × 10 double], diffusion signal measured at b = 0 (ten times). Usually, we compute S0(v) by averaging over the ten available values.
• fe.life.bvecs: [Nθ × 3 double], each row indicates the gradient 3D di- rection.
• fe.life.bvals: [Nθ × 1 double], each row indicates the used b-value for each gradient.
• fe.life.bvecsindices: [Nθ × 1 int], indices for measurements with non- zero b-value.
• fe.life.imagedim: [Nx , Ny , Nz , Nm ], size of the input 4D dMRI dataset where (Nx,Ny,Nz) are the the sizes in each coordinate, x, y and z, respec- tivelly; and Nm correspond to the number of measurements with b = 0 and b ̸= 0.
• fe.life.voxel2FNpair: not used.
• fe.life.modelTensor: [λ1 , λ2 , λ3 ], parameters of the Diffusion Tensor (DT) model used to generate the Dictionary of diffusion kernels.
• fe.life.fit: this field contains the results of applying the LiFE method, see description below.
fe.life.fit fields
• fe.life.fit.randState: saved by the optimization algorithm.
• fe.life.fit.results: saved by the optimization algorithm.
• fe.life.fit.weights: [Nf × 1 double], fascicles weights obtained as a result of the optimization.
• fe.life.fit.params.fitMethod: [text], name of used optimization method, default is “bbnls”.
fe.fg fields
• fe.fg.name: [text], name of Fiber Group (connectome).
• fe.fg.colorRgb: [R, G, B], color specification for visualization.
• fe.fg.thickness: [double], thickness specificattion for visualization.
• fe.fg.visible: [binary], parameter for visualization.
• fe.fg.seeds: Seeds used by MRTRIX software to generate the connectome.
• fe.fg.seedRadius: MRTRIX parameter.
• fe.fg.seedVoxelOffsets: MRTRIX parameter.
• fe.fg.params: additional MRTRIX parameters.
• fe.fg.fibers: {Nf ×1 cell}, set of fascicles (streamlines), generated with a tractography algorithm. Each cell contains the x,y,z coordinates a list of 3D points describing the trajectory of a single fascicle.
fe.roi fields
• fe.roi.name: [text], name of ROI.
• fe.roi.color: [R, G, B], color specification for visualization.
• fe.roi.coords: [Nv ×3 double], voxel coordinates, each row specifies the spatial location of a single voxel.
• fe.roi.visible: [binary], parameter for visualization.
• fe.roi.mesh: not used.
• fe.roi.dirty: parameter for visualization
• fe.roi.query_id: not used.
Notes
This software is currently compatible with Matlab 2020b or higher and only with Mac OSX and Linux (Ubuntu/Fedora).
-
python内部的字符串一般都是 Unicode编码。代码中字符串的默认编码与代码文件本身的编码是一致的。所以要做一些编码转换通常是要以Unicode作为中间编码进行转换的,即先将其他编码的字符串解码(decode)成 Unicode,再从 Unicode编码(encode)成另一种编码。 decode 的作用是将其他编码的字符串转换成 Unicode 编码,eg name.decode(“GB2
-
从英文意思上看,encode和decode分别指编码和解码。在python中,Unicode类型是作为编码的基础类型,即: decode encode str ---------> str(Unicode) ---------> str 1 2 >>> u = '中文' # 指定字符串类型对象u >>> str1 = u
-
问题内容: 我有一个实现了Serializable的TouchPoint类,因为它包含位图,所以我为该类编写了writeObject和readObject: 问题是我得到了 SkImageDecoder :: Factory返回null 那么我该如何解决。我知道可能的解决方案是将writeObject()更改为 但是这种方法要慢10倍以上。 copyPixelsToBuffer〜14ms用于写入图
-
java.lang.NoSuchMethodError:Java EE应用程序中的org.apache.commons.codec.binary.Base64.encodeBase64String()
问题内容: 我正在开发需要Base64编码/解码的Java EE应用程序 所以我在应用程序的文件夹中添加并使用了 在Java文件中。 在编译期间,当我输入时,它表明method可用。但是在运行时,它将引发如下异常: 我在构建路径中有JAR,但我仍然不明白为什么它会引发上述错误。 问题答案: 该方法在Commons Codec 1.4中引入。此异常表示您在Web应用程序的运行时类路径中的其他位置具有
-
问题内容: 我一直在尝试各种Java代码,试图提出一些将对包含引号,空格和“奇异” Unicode字符的字符串进行编码的东西,并产生与JavaScript的encodeURIComponent函数相同的输出。 我的酷刑测试字符串是: “ A” B±“ 如果我在Firebug中输入以下JavaScript语句: —然后我得到: 这是我的小测试Java程序: —该程序输出: 靠近,但没有雪茄!使用Ja
-
问题内容: 编码要发送到Web服务器的查询字符串时-您何时使用以及何时使用或: 使用转义: 要么 使用encodeURI()/ encodeURIComponent() 问题答案: escape() 不要使用它! 在B.2.1.2节中定义了转义,并且附件B的引言中指出: …本附件中指定的所有语言功能和行为均具有一个或多个不良特征,在没有遗留用法的情况下,将从本规范中删除。… …程序员不应该使用或编
-
本文向大家介绍JavaScript实现的encode64加密算法实例分析,包括了JavaScript实现的encode64加密算法实例分析的使用技巧和注意事项,需要的朋友参考一下 本文实例讲述了JavaScript实现的encode64加密算法。分享给大家供大家参考。具体如下: 这段JavaScript代码可实现encode64加密算法,速度还是相当不错的。 希望本文所述对大家的javascrip
-
本文向大家介绍JavaScript使用encodeURI()和decodeURI()获取字符串值的方法,包括了JavaScript使用encodeURI()和decodeURI()获取字符串值的方法的使用技巧和注意事项,需要的朋友参考一下 本文实例讲述了JavaScript使用encodeURI()和decodeURI()获取字符串值的方法。分享给大家供大家参考。具体如下: JavaScript使
-
本文向大家介绍详解Python解决抓取内容乱码问题(decode和encode解码),包括了详解Python解决抓取内容乱码问题(decode和encode解码)的使用技巧和注意事项,需要的朋友参考一下 一、乱码问题描述 经常在爬虫或者一些操作的时候,经常会出现中文乱码等问题,如下 原因是源网页编码和爬取下来后的编码格式不一致 二、利用encode与decode解决乱码问题 字符串在Python
-
本文向大家介绍js中字符串编码函数escape()、encodeURI()、encodeURIComponent()区别详解,包括了js中字符串编码函数escape()、encodeURI()、encodeURIComponent()区别详解的使用技巧和注意事项,需要的朋友参考一下 JavaScript中有三个可以对字符串编码的函数,分别是: escape,encodeURI,encodeURIC